NoteSeQ can assess Cas9, Cas12, and adenine/cytosine base and prime editors, while producing variant-aware analysis of thousands of genomes simultaneously to detect a broad spectrum of off-target sites. These nominations are then orthogonally validated, adhering to the FDA’s draft guidance. Target cells or tissues are then tested to confirm which of the nominated sites are altered after therapeutic gene editing
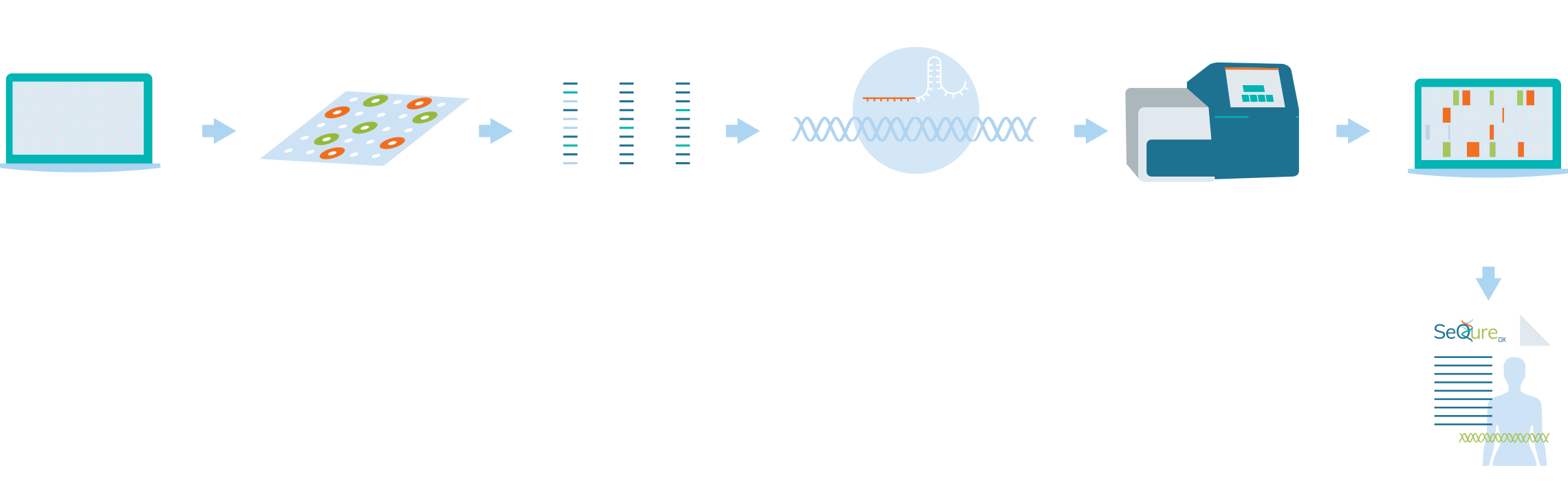
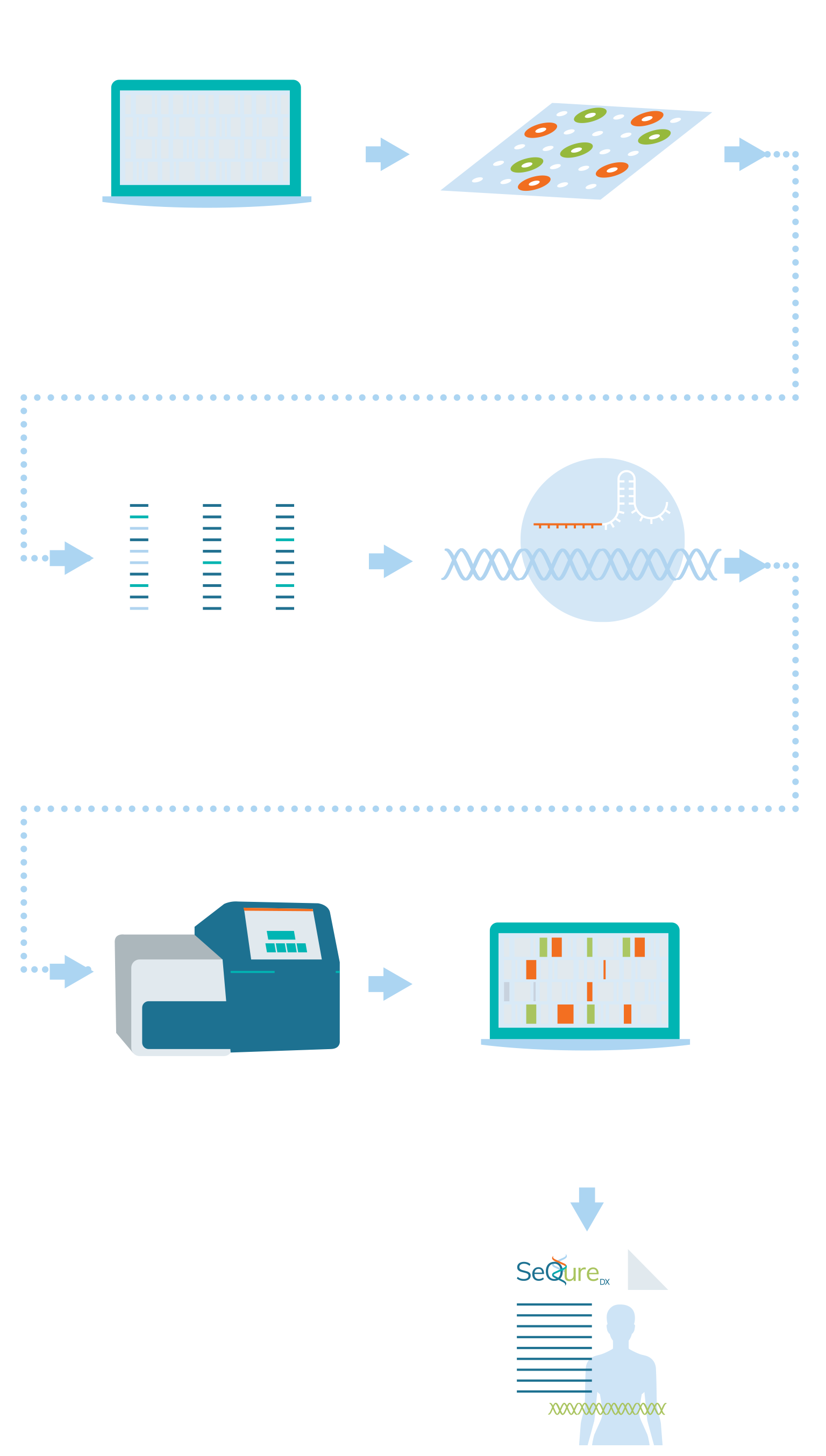
Workflow
SeQure Dx has a highly scalable and gene editor agnostic platform that provides comprehensive off-target editing results.
At the core of our NoteSeQ platform is the ONE-seq assay. Using our comprehensive bioinformatic platform to produce a computational enrichment of all potential off-target edits in the reference genome, all potential genetic edited sites are synthesized in oligos in a uniform library. These oligonucleotides are in vitro edited using the editing enzyme of interest and the edited sites are sequenced. Post-sequence bioinformatic analysis creates a report that is assessed for the biologic impact of the genetic editing for an individual, ethnic group, or specific cell line.